|
# coding: utf-8
"""
This script can help you to summary the plink2 report file
"""
import os
import re
work_path = r"C:\Users\86181\Documents\pLink\HOIPSDApLink_task_2022.11.19.16.30.07\reports"
spec_cutoff = 1 # spectra number cut-off
Best_evalue_cutoff = 2 # 交联位点对层次最好的e-value cutoff
E_value_cutoff_SpecLvl = 2 # 谱图层次的e-value cutoff
################Don't change the following lines###############
if work_path:
reports_path = work_path
else:
reports_path = os.getcwd()
def count_keyIndic(ele, ele_dic):
if ele not in ele_dic:
ele_dic[ele] = 1
else:
ele_dic[ele] += 1
#根据交联位点信息和交联肽段信息判断inter还是intra
def judgeHomoHetro(linked_site, pepXL):
pos_list = re.findall("\((\d*)\)", linked_site)
position1 = pos_list[0]
position2 = pos_list[1]
p = linked_site.find(")-")
m = linked_site.find("(" + position1 + ")-")
n = linked_site.find("(" + position2 + ")", p)
protein1 = linked_site[:m]
protein2 = linked_site[p + 2:n]
if protein1 != protein2:
return "Inter"
else:
linkPosPep1, linkPosPep2 = re.findall("\((\d*)\)", pepXL)
p1 = pepXL.find(")-")
m1 = pepXL.find("(" + linkPosPep1 + ")")
n1 = pepXL.find("(" + linkPosPep2 + ")", p1)
pep1 = pepXL[:m1]
pep2 = pepXL[p1 + 2:n1]
deltaList = [
int(position1) - int(linkPosPep1),
int(position2) - int(linkPosPep2)
]
pep1IDX = [1 + deltaList[0], len(pep1) + deltaList[0]]
pep2IDX = [1 + deltaList[1], len(pep2) + deltaList[1]]
if pep1IDX[1] < pep2IDX[0] or pep1IDX[0] > pep2IDX[1]:
return "Intra"
else:
return "Inter"
# 位点处理,将位点对里面的反库蛋白和污染蛋白的交联对剔除
def site_list_process(site_list, pepXLlist=["WFC(2)-XSV(2)"]):
i = 0
while i < len(site_list):
if "REVERSE" in site_list[i] or "gi|CON" in site_list[i]:
site_list.remove(site_list[i])
else:
i += 1
if site_list == "":
return "", None
else:
link_type_list = []
for i in range(len(site_list)):
boolInter = False
for j in range(len(pepXLlist)):
if judgeHomoHetro(site_list[i], pepXLlist[j]) == "Inter":
boolInter = True
break
else:
continue
if boolInter:
link_type_list.append("Inter")
else:
link_type_list.append("Intra")
linkType = "/".join(link_type_list)
site = "/".join(site_list)
return site, linkType
# 获取报告文件的名称,读取上级文件夹下的.plink文件查找交联剂和酶的名称,若找不到则返回“pLink_summary.csv”
def get_report_file_name():
path_d = os.path.dirname(os.getcwd())
try:
file_list = os.listdir(path_d)
for fl in file_list:
if fl.endswith("plink"):
para = open(os.path.join(path_d, fl)).readlines()
for line in para:
if line.startswith("spec_title"):
spec_title = line.split("=")[1].strip()
if line.startswith("enzyme_name"):
enzyme = line.split("=")[1].strip()
if line.startswith("linker1"):
linker = line.split("=")[1].strip()
report_file_name = "_".join([spec_title, linker, enzyme, "v5.csv"])
return report_file_name
return "pLink_summary.csv"
except:
return "pLink_summary.csv"
# 根据报告文件获取所有raw文件的名称
def get_crosslink_site_info(site_table):
raw_name_list = []
for line in site_table[2:]:
line_list = line.rstrip("\n").split(",")
if line_list[0] == "":
raw_name = line_list[2].split(".")[0]
if raw_name not in raw_name_list:
raw_name_list.append(raw_name)
raw_name_list.sort()
return raw_name_list
#计算openedfl的某一列k的和
def cal_sumOfOneColumn(openedfl, k_column):
f = openedfl
k = k_column
sumNum = 0
for line in f[1:]:
lineList = line.rstrip("\n").split(",")
val = lineList[k]
if val:
sumNum += int(lineList[k])
return sumNum
#计算openedfl某一列k的取值范围
def cal_numRange(openedfl, k_column):
f = openedfl
k = k_column
valList = []
for line in f[1:]:
lineList = line.rstrip("\n").split(",")
if lineList[k]:
valList.append(float(lineList[k]))
valList.sort()
return "{0:.1e}~{1:.1e}".format(valList[0], valList[-1])
def count_peptides(f):
num_pep = 0
for line in f[1:]:
peps = line.split(",")[4]
num_pep += peps.count(";") + 1
return num_pep
def statistic_report_file():
report_file_name = get_report_file_name()
c = open(report_file_name, 'a')
f = open(report_file_name, 'r').readlines()
col_num = len(f[0].split(","))
stat_list = [""]*col_num
total_colom = len(f[0].split(","))
ttl_sites_num = len(f) - 1
intra_num = 0
for i in range(1, len(f)):
if f[i].strip("\n").split(",")[5] == "Intra":
intra_num += 1
stat_list[5] = round(intra_num / ttl_sites_num, 2)
stat_list[0] = ttl_sites_num
stat_list[1] = cal_sumOfOneColumn(f, 1)
stat_list[4] = count_peptides(f)
column_sub_dic = {}
for k in [6, 8]:
for j in range(k, total_colom, 3):
stat_list[j] = cal_sumOfOneColumn(f, j)
for j in range(7, total_colom, 3):
stat_list[j] = cal_numRange(f, j)
c.write(",".join([str(ele) if type(ele) != str else ele \
for ele in stat_list]) + "\n\n")
c.write("Raw_Name,# of Pep,# of Spec,e-value range\n")
for i in range(6, col_num, 3):
raw_name_list = f[0].split(",")[i].split("_")[:-1]
raw_name = "_".join(raw_name_list)
wlist = [stat_list[i+2], stat_list[i], stat_list[i+1]]
wlist.insert(0, raw_name)
c.write(",".join([str(ele) for ele in wlist])+"\n")
c.close()
def splitResult(openedfl, raw_name_list, spec_cutoff, Best_evalue_cutoff, E_value_cutoff_SpecLvl=2):
finalList = []
f = openedfl
n = 2
while n < len(f):
spec_dic = {}
pep_dic = {}
evalue_dic = {}
line_list = f[n].rstrip("\n").split(",")
if line_list[0].isdigit():
site_list = [line_list[1]]
p = n + 1
else:
print("文件格式错误,line%d"%n)
while p < len(f):
line_list = f[p].rstrip("\n").split(",")
if line_list[0] == "" and line_list[1].isdigit():
break
else:
if line_list[0] == "SameSet":
site_list.append(line_list[1])
p += 1
site = site_list_process(site_list)[0]
if site == "":
while p < len(f):
if f[p].rstrip("\n").split(",")[0].isdigit():
break
else:
p += 1
else:
bestSVMscore = f[p].rstrip("\n").split(",")[9]
pep_std_list = [] # f[p].rstrip("\n").split(",")[5]
while p < len(f): # and f[p].rstrip("\n").split(",")[0] == "":
line_list = f[p].rstrip("\n").split(",")
if line_list[0].isdigit():
break
else:
pep_std = line_list[5]
if pep_std not in pep_std_list:
pep_std_list.append(pep_std)
evalue = float(line_list[8])
if evalue < E_value_cutoff_SpecLvl:
pep = line_list[5]
raw_name = line_list[2][:line_list[2].find(".")]
if raw_name not in spec_dic:
spec_dic[raw_name] = 1
else:
spec_dic[raw_name] += 1
if raw_name not in pep_dic:
pep_dic[raw_name] = [pep]
else:
if pep not in pep_dic[raw_name]:
pep_dic[raw_name].append(pep)
if raw_name not in evalue_dic:
evalue_dic[raw_name] = evalue
else:
if evalue < evalue_dic[raw_name]:
evalue_dic[raw_name] = evalue
p += 1
totalPep = ";".join(pep_std_list)
link_type = site_list_process(site_list, pep_std_list)[1]
total_spec = 0
min_evalue = 1
for key in evalue_dic:
total_spec += spec_dic[key]
if evalue_dic[key] < min_evalue:
min_evalue = evalue_dic[key]
else:
continue
if total_spec >= spec_cutoff and min_evalue < Best_evalue_cutoff:
rep_list = [site, total_spec, min_evalue,\
bestSVMscore, totalPep, link_type]
for raw in raw_name_list:
if raw not in spec_dic:
SEP = ["", "", ""]
else:
SEP = [
spec_dic[raw], evalue_dic[raw],
len(pep_dic[raw])
]
rep_list.extend(SEP)
finalList.append(",".join([str(ele) for ele in rep_list]))
n = p
finalList = sorted(finalList,
key=lambda x: int(x.split(",")[1]),
reverse=True)
return finalList
def find_xlPeptides_File(reports_path):
for fl in os.listdir(reports_path):
if fl.endswith("cross-linked_sites.csv"):
return fl
return ""
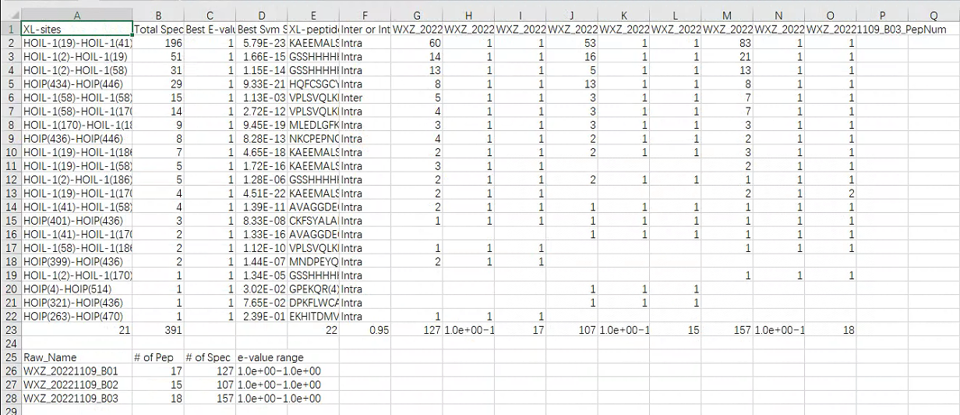
def write2report(raw_name_list, final_list):
report_file_name = get_report_file_name()
b = open(report_file_name, 'w')
col = ["XL-sites", "Total Spec", \
"Best E-value", "Best Svm Score", "XL-peptide", "Inter or Intra Molecular"
]
for name in raw_name_list:
col.append(name + "_SpecNum")
col.append(name + "_E-value")
col.append(name + "_PepNum")
b.write(','.join(col)+"\n")
for line in final_list:
b.write(line + "\n")
b.close()
def main_flow(reports_path, spec_cutoff, Best_evalue_cutoff):
os.chdir(reports_path)
xlsitesfl = find_xlPeptides_File(reports_path)
if xlsitesfl == "":
print("Please check your file")
else:
f = open(xlsitesfl).readlines()
raw_name_list = get_crosslink_site_info(f)
finalList = splitResult(f, raw_name_list, spec_cutoff, Best_evalue_cutoff)
write2report(raw_name_list, finalList)
print("The task is finished")
statistic_report_file()
if __name__ == "__main__":
main_flow(reports_path, spec_cutoff, Best_evalue_cutoff) |
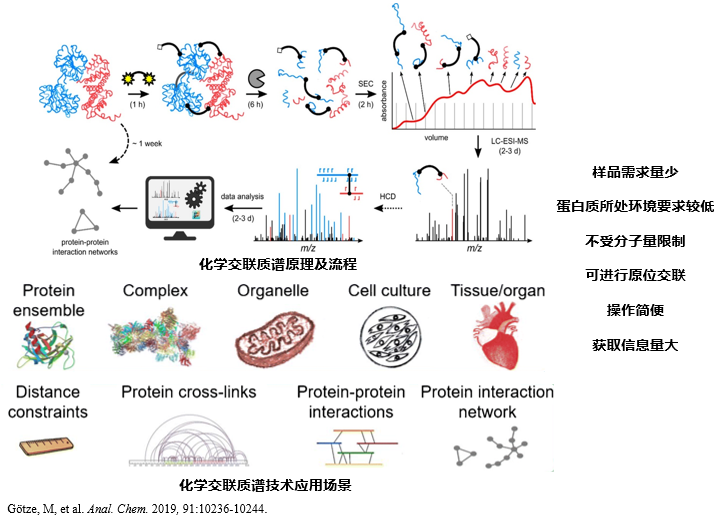
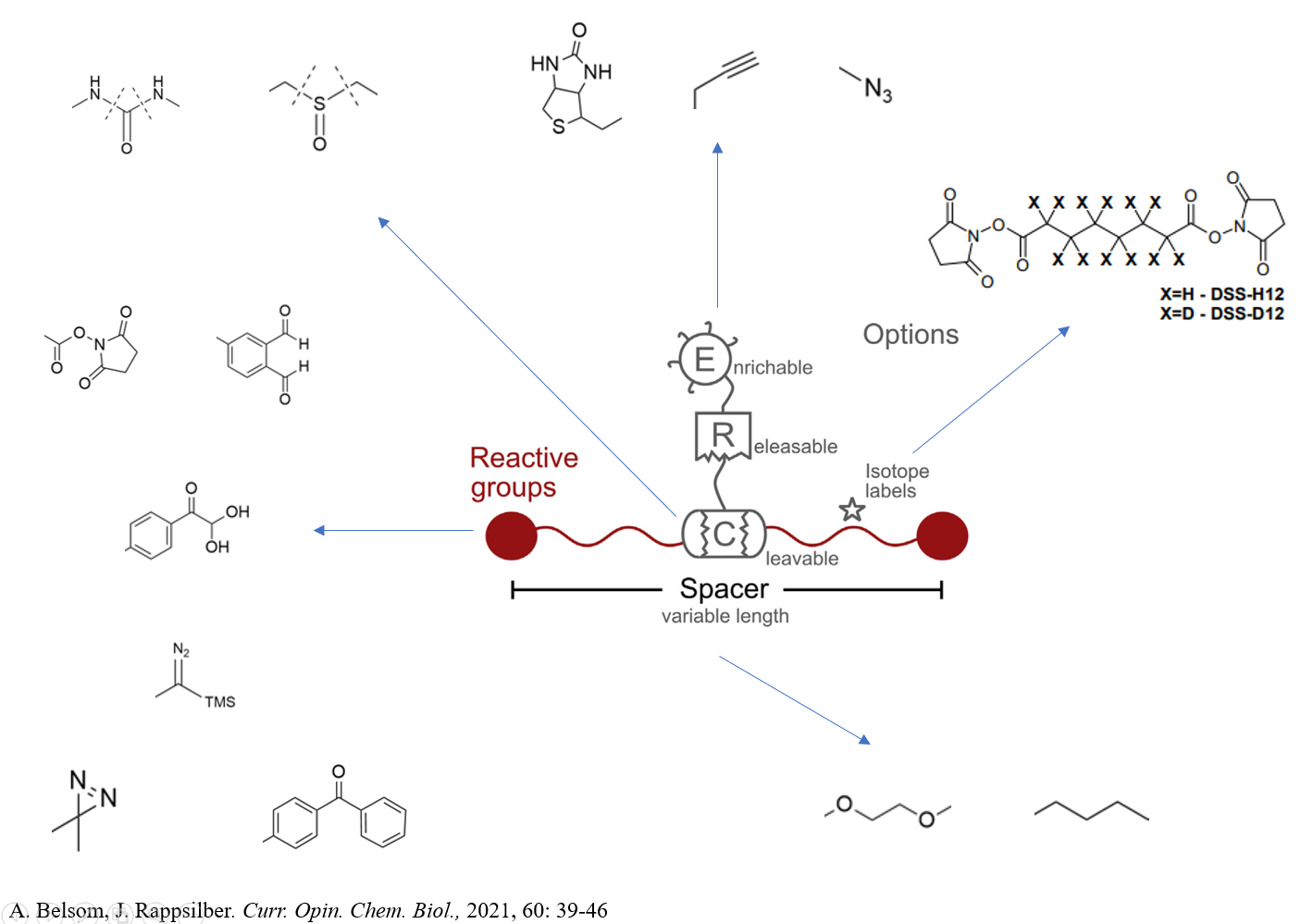
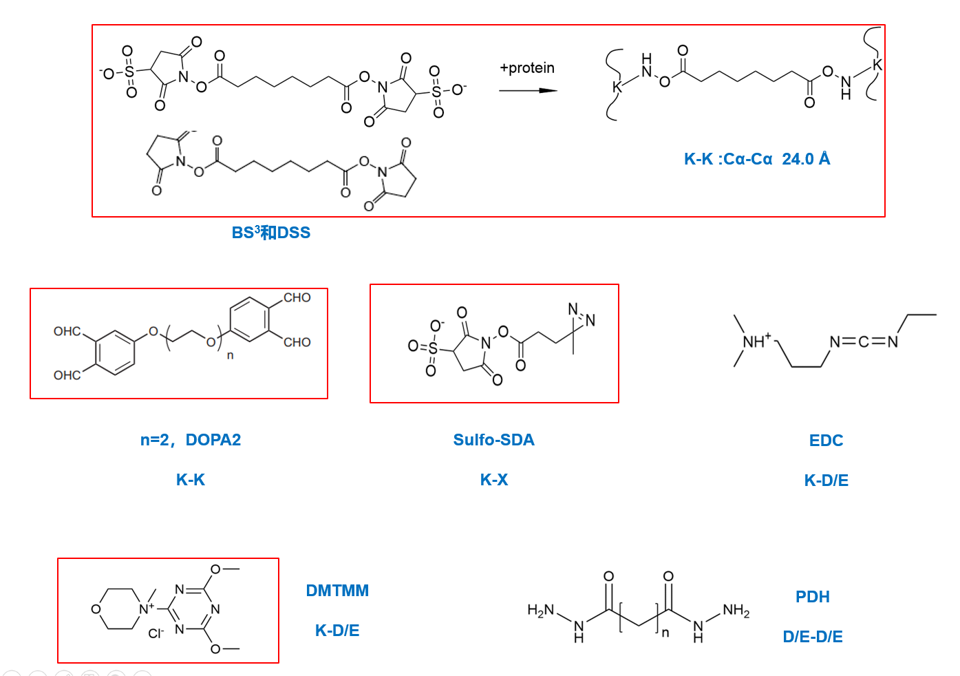
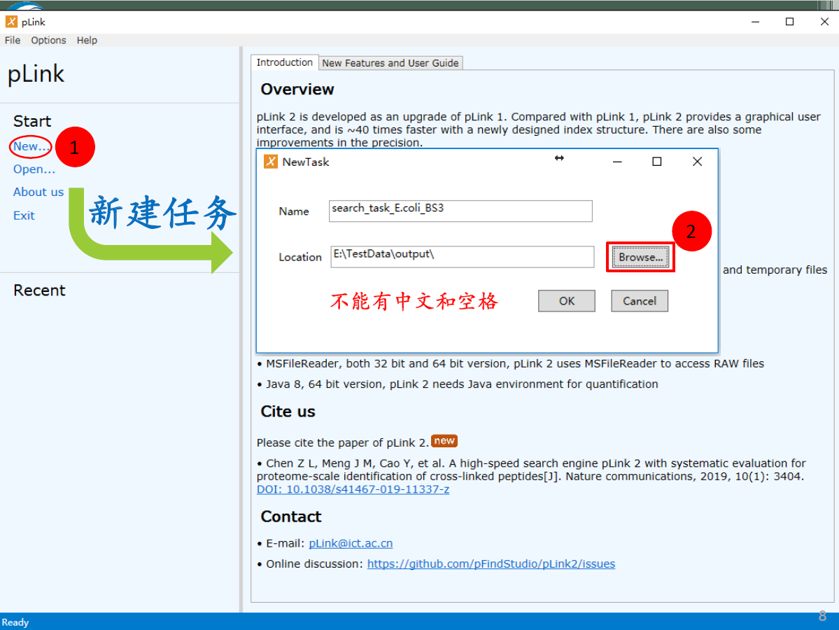
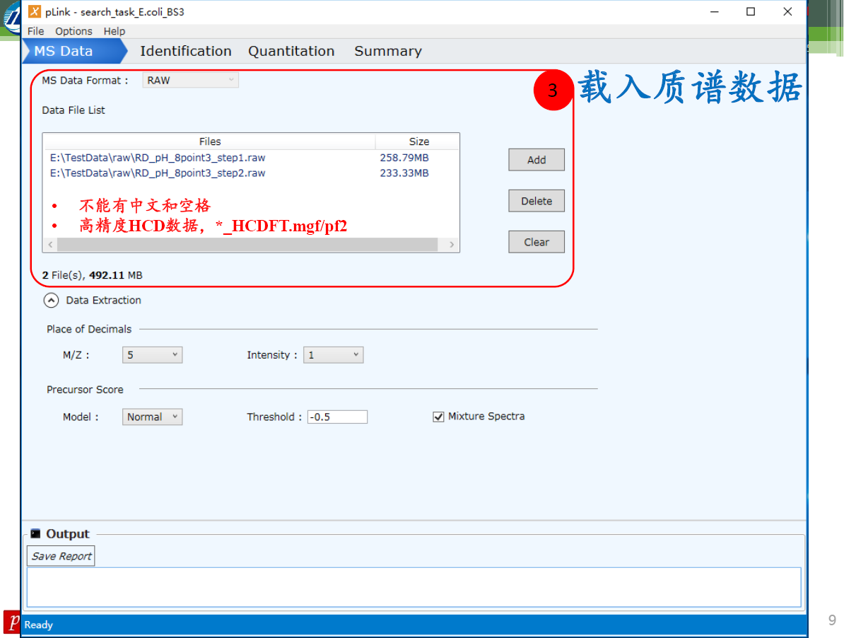
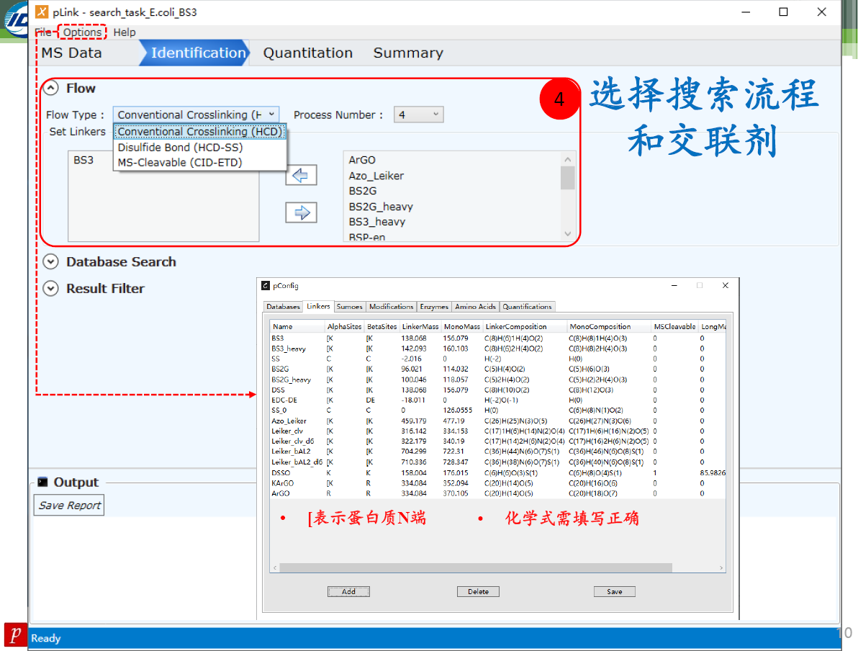
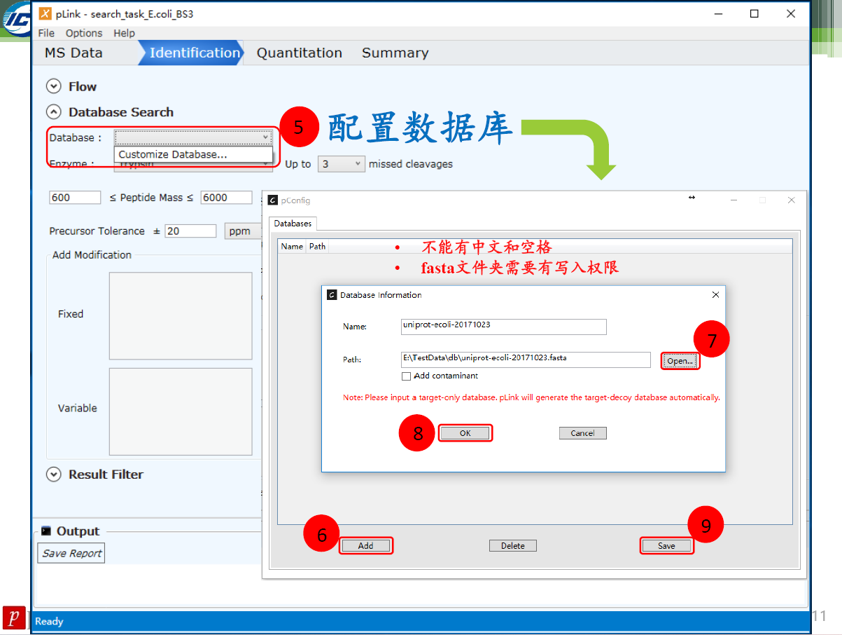
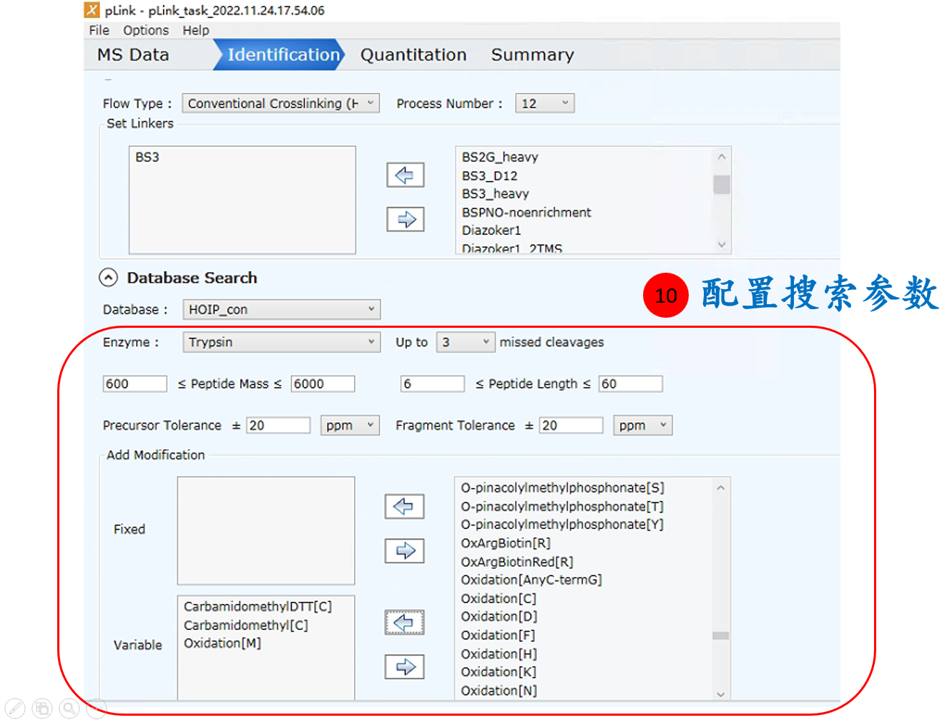
![]()